小豆开花时间全基因组关联分析

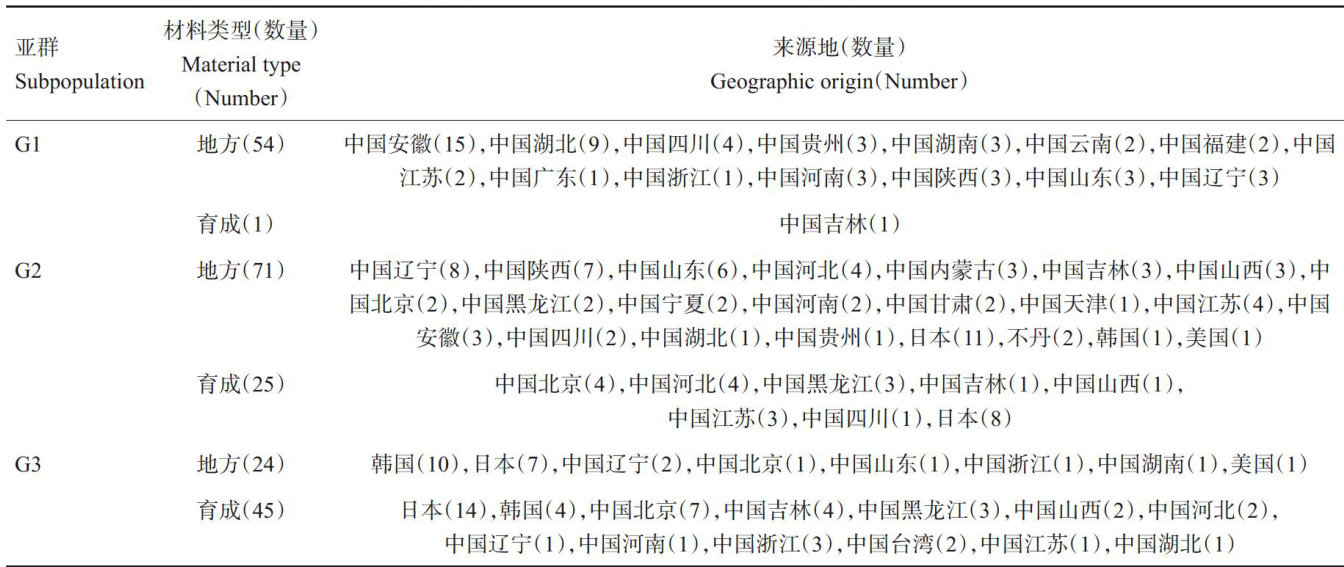
打开文本图片集
Abstract: Identifying genetic loci associated with flowering time in adzuki bean (Vigna angularis L.) is critical for breeding and agricultural production.This study investigated flowering time variations across 220 cultivated adzuki beanaccessons through biennial field trialsconducted in Ma'anshan city,Anhui province, China. Population structure analysis was performed using SNPs obtained from whole-genome resequencing, followed by genome-wide association analysis (GWAS)of flowering time employing a mixed linear model. The population structure analysis revealed three distinct subpopulations (G1,G2,G3),which correlated with geographical origins and variety type. Substantial phenotypic variation in flowering time was observed across adzuki bean accessions,ranging from 40 to 81 days,with significant diffrences observed among subpopulations. GWAS identified eight quantitative trait loci (QTLs) for flowering time distributed across chromosomes 2,3, 4,5,10,and 11.Functional annotation of genes within these QTL intervals prioritized nine candidate genes potentially regulating flowering time.Haplotype analysiswasperformed on fourcandidate genes, Vigan02g270300 (Clp1 family protein in precursor mRNA splicing complex II),Vigan10g218300 (CDF transcription factor),Viganllg07950o(CONSTANS-like zinc finger protein),and Viganllg080900 (vernalization insensitive protein VIL1),whose homologous genes in model plants directly regulate flowering time.Among these,the haplotype Hap2 of Vigan10g218300 was associated with early flowering.These findings provide a theoretical basis for further analyzing the genetic mechanism of flowering regulation in adzuki bean and breeding cultivars with optimized growth periods.
Key words: adzuki bean; flowering time ; genome-wide association analysis ;candidate genes ;haplotype
小豆(VignaangularisL.)隶属豆科豇豆属,是一种兼具营养与保健功能的杂粮经济作物[1],其生育期短、适应性广,可与其他作物轮作倒茬和间作套种,在调整种植业结构和耕作制度、促进资源节约型农业发展、实现固氮环保等方面有着重要作用[2-3]。(剩余18590字)