基于Ilumina数据的莱茵衣藻聚腺苷酸化生物信息学研究

打开文本图片集
中图分类号:Q756 文献标识码:A 文章编号:1674-0033(2025)04-0052-06
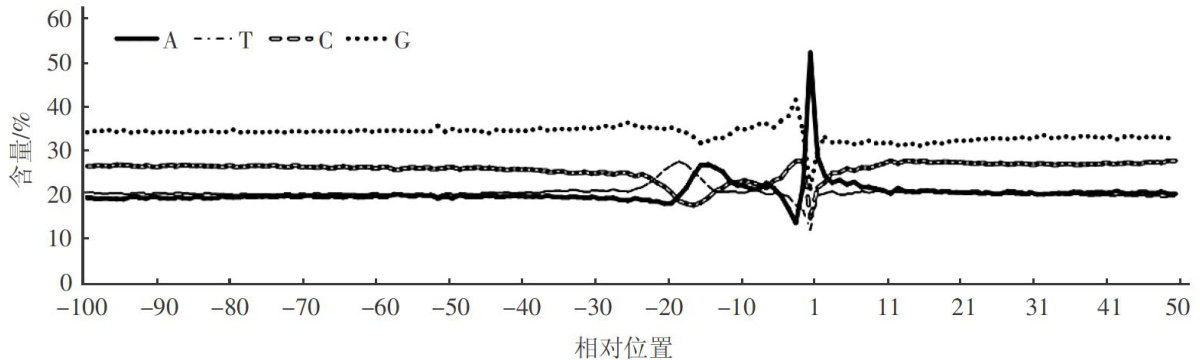
Abstract:Inorder to investigate thecharacteristicsof polyadenalation (poly(A))and alternative polyadenalation (APA) in the genome of Chlamydomonas reinhardtii, 23 535 153 RNA-Seq reads with poly(A) tails were collected from the high throughput illuminadata,and then 256771 unique poly(A)sites without redundant and 97 479 PACs (poly(A) clusters) were collected based on the reads. It is illuminated that 55.67% and 28.48% (20 unique poly(A) sites were detected in 3′ -UTR and intergenic regions,respectively. Single nucleotide profile showed adenine (A) peak (52.20 % )around the poly(A) sites,and guanine (G, 33.72% averagely) was obviously higher than other three, UGUAA (19.76 % ,Z-score=32.88) was a dominant poly(A) signal in NUE region. The APA genes were up to 67.78% in C.reinhardti.Furthermore,GO analysis demonstrated that the genes mainly involved in molecular function and biological proces,and revealed the genes participated in the formation of multiple protein isoforms by changing receptor activity and ncRNA metabolic process.
Key words:Poly(A) signals; alternative polyadenylation; Ilumina data; GO analysis;Chlamydomonas reinhardtii
信使RNA(mRNA) 3′ 端的形成包括剪切和多聚腺苷酸化(polyadenylation,poly(A))过程,该过程是真核生物转录后加工的关键步骤。(剩余7711字)